在ggplot 中,我们可以使用geom_bar 来画柱状图。
但默认下,柱状图并不需要定义y,我们只需要制定相应的x 或进一步的分组(fill 等),就会对数据进行计数。
但在某些情况下,我们的数据框可能非常大,这时候就可以自己进行计数,然后告诉ggplot 即可。
有两种指定y 的方式:
geom_colgeom_bar(stat = 'identity')
它们的结果都是一样的。
那么该如何分组计数呢?
也非常简单,tidyverse 套件提供了group_by 分组以及summarise 函数,使用n() 计算。
或者直接基础的table 搞定:
## count variant in each sampletmp1 <- table(mutation_number_order$name, mutation_number_order$Variant_Classification)tmp1 <- as.data.frame(tmp1)colnames(tmp1)[1:2] <- c("name", "Variant_Classification")> head(tmp1)name Variant_Classification Freq1 S110011502DT Frame_Shift_Del 382 S110011501DT Frame_Shift_Del 413 S110020203DT Frame_Shift_Del 364 S110030206DT Frame_Shift_Del 435 S110030801DT Frame_Shift_Del 506 S110020201DT Frame_Shift_Del 49
合并到原表格中,直接画就完事了:
## count variant in each sampletmp1 <- table(mutation_number_order$name, mutation_number_order$Variant_Classification)tmp1 <- as.data.frame(tmp1)colnames(tmp1)[1:2] <- c("name", "Variant_Classification")head(tmp1)tmp3 <- merge(mutation_number_order, tmp1, by = c("name", "Variant_Classification"))mutation_number_final <- unique(tmp3)colnames(mutation_number_final) <- c("Tumor_Sample_Barcode","Variant_Classification","Clinical_Type","Total_Counts","Counts")max_counts <- max(mutation_number_final$Total_Counts)# counts plotp1 <- ggplot(data = mutation_number_final) +geom_col(mapping = aes(x = Tumor_Sample_Barcode, y = Counts, fill = Variant_Classification), position = "stack") +barplot_theme + labs(x = NULL, size = 14) + scale_y_continuous(expand=c(0,0)) +coord_cartesian(ylim = c(0, max_counts + 100))(p1 <- p1 + labs(y = "Mutation Counts") + scale_x_discrete(expand = expansion(mult = c(0.03,0.05))))
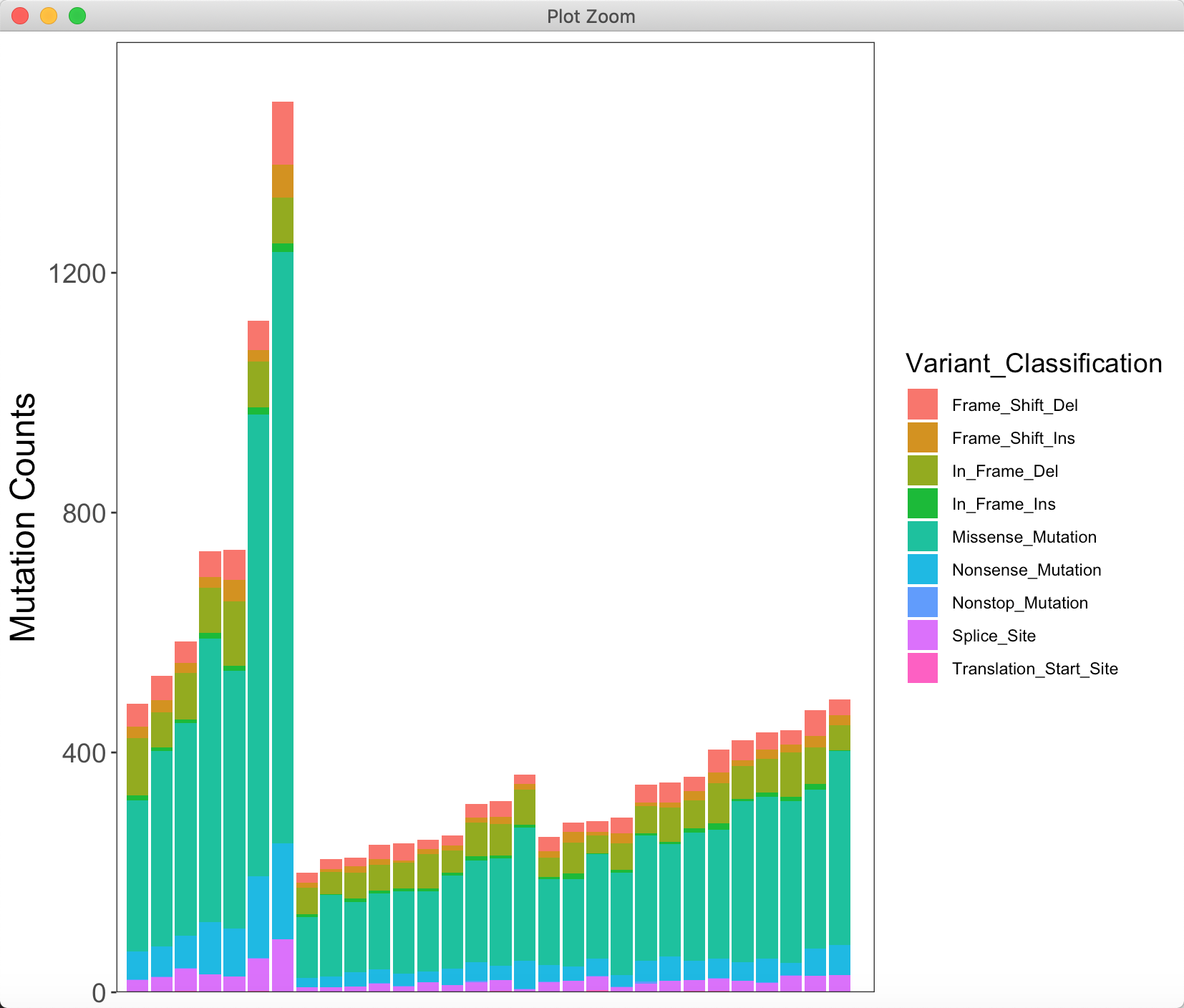
y 也就是table 最后算出来的各组的数值,fill 分组的变量,再stack 也就堆积到一起啦。

